Introduction
The phyloviz app contains 3 types of pages:
- The virus Family pages
- Individual virus pages
- The toolbox page
Depending on the type of page, the content is collected and shown differently.
Data loading
At the basis are a number of data files. The primary ones are:
src/families.yaml: This file contains the different families that are represented in the app. This is just a simple list.src/toolbox.yaml: The content of the toolbox, including meta information.
We use YAML format here because it’s easy for humans to write and interpret. Internally, we use the JSON representation and thus we neekkkkkkd to convert them to the JSON format. That’s what we use yq for:
yq -M -o=json src/toolbox.yaml > toolbox/toolbox.json
yq -M -o=json src/families.yaml > families.jsonThese files provide general information about the toolbox and the different families that are suppored. Once they are parsed and read, the per-familiy Excel annotation files are read.
Next, per-family information is read in. For every family (say virfam) in families.json, the following data files are loaded and parsed:
virfam/tree.newick: this contains the phylogenetic tree information.virfam/family.xlsxcontains the annotations for the different virus strains of interest.
While parsing the different data sources, a translation is performed so that tools in the tooblox can be referred to by means of a short identifier rather than their longer names. These data files are loaded for every page within the app. This is done in the _js/_ojs_data.qmd.
Virus family page
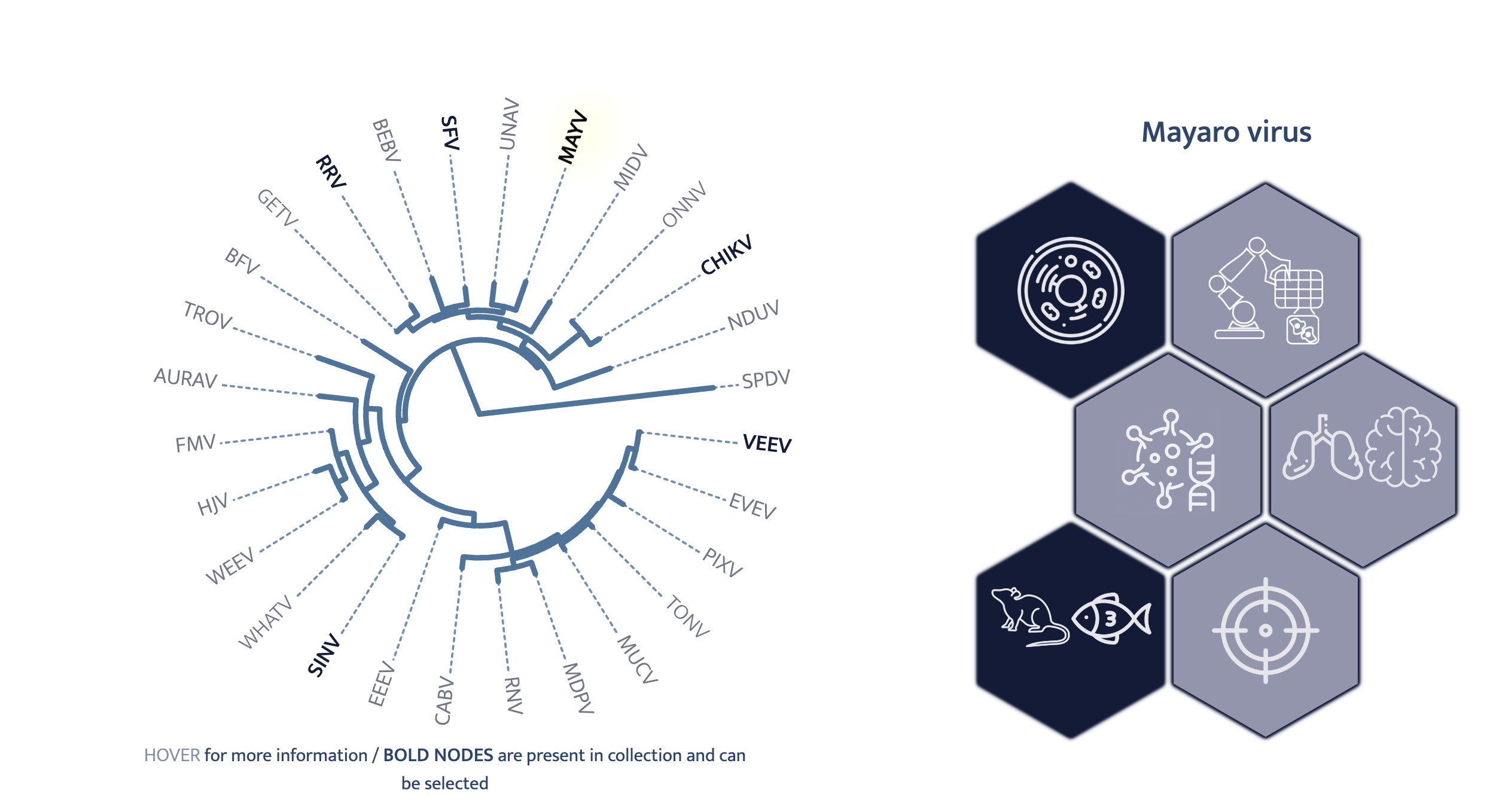
As an illustration, this is the alphaviruses/index.qmd file:
---
...
params:
family: alphaviruses
---
```{r}
#| echo: false
ojs_define(virusFamily = params$family)
```
{{< include /_js/_ojs_data.qmd >}}
{{< include /_js/_ojs_family.qmd >}}
{{< include /_js/family-breadcrumb.qmd >}}
{{< include /_js/family.qmd >}}
{{< include _details.qmd >}}A virus family first loads all the data for all families (via the _js/_ojs_data.qmd file discussed earlier). As a second step, this data is filtered for the virus at hand, say virfam. This is done by means of a parameter that is set in the family page .qmd file as can be seen above.
So based on the family parameter, the data is filtered and the appropriate phylotree data and virus strain annotations are extracted1.
The utility file _js/_ojs_family.qmd contains a number of helper functions and transformations that deal with the filtering and parsing of the data structures for one family.
Tree and toolbox rendering
The effective rendering of the tree and the toolbox is done in _js/family.qmd and that file is included in every virus family index.qmd file as can be seen as well.
The code should be self-explanatory, so we’re not going to discuss it in detail here.
Details
The last include in the virus family page is _details.qmd. Please note that this is not a shared file as it resides in the same directory as the family information itself.
This is a sample from alphaviruses/_details.qmd:
:::: {.panel-tabset #vb-tabset}
## MAYV
{{< include _MAYV.qmd >}}
:::{ style="text-align:center;font-weight:500" }
[open virus page](/alphaviruses/MAYV.qmd) / [reload family page](/alphaviruses/)
:::
## SFV
{{< include _SFV.qmd >}}
:::{ style="text-align:center;font-weight:500" }
[open virus page](/alphaviruses/SFV.qmd) / [reload family page](/alphaviruses/)
:::Clearly, this is not a generic file in that it contains explicit references to the viruses for the alphavirus family2.
The last we want to do is type such a virus list ourselves. The probability of errors is huge and it’s the most boring task there is.
We generate the <family>/_details.qmd files based on the annotations present in the family XLSX file using a script: _js/update_family.js. This script is run using node.js:
node _js/update_family.js -i alphavirusesThis will read all the necessary data files and generate the _details.qmd file (as well as a bunch of other files we will come back to later).
A convenience function has been created that generates all these files for ALL virus families: _js/update_families.js.
Please note:
these tools will not overwrite anything that is already present. This is a failsafe. In ordert to explicitly overwrite the existing files, one can either first remove them or use the
-oargument to the tools.The virus-specific content that is loaded from the
_details.qmdpage are in itself files that start with_and so are not rendered.
Let us take a loook at for instance _MAYV.qmd:
::: {.panel-tabset}
## Symptoms
::: {.virus-figure}
{{< include _MAYV-symptoms-fig.qmd >}}
:::
::: {.virus-text}
{{< include _MAYV-symptoms.qmd >}}
:::
## Transmission
::: {.virus-figure}
{{< include _MAYV-transmission-fig.qmd >}}
:::
::: {.virus-text}
{{< include _MAYV-transmission.qmd >}}
:::
## Medical relevance
...
## Toolbox
...
:::We notice a pattern, this file, although auto-generated by the update_family.js script in itself only refers to other files in turn. For example, _MAYV-symptoms.qmd:
More than 50% of the infected patients develop a chronic, long-term disease, characterized by a debilitating arthralgia that can persist for weeks to months.
Death from MAYV is rare.or _MAYV-symptoms-fig:
These are (for the most part) all simple (markdown) text files. They provide the content for the virus details panes.
Please note that all this content is loaded in the tabs below the phylotree and toolbox visualization.
Virus page
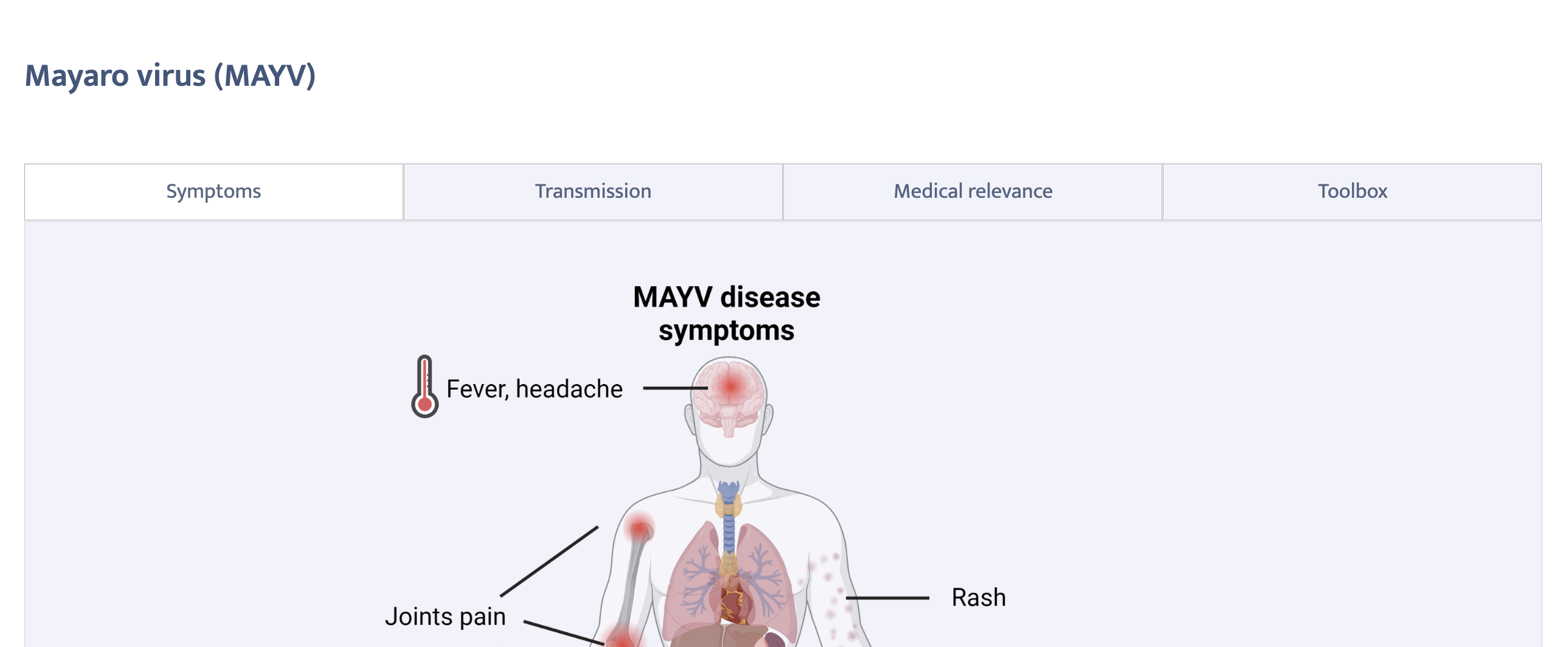
When we click on a specific virus, we are taken to the virus page. Let’s consider the example of alphaviruses/MAYV.qmd again:
---
params:
family: alphaviruses
virus: Mayaro virus (MAYV)
---
```{r}
#| echo: false
ojs_define(virusFamily = params$family)
ojs_define(virus = params$virus)
```
{{< include /_js/virus-breadcrumb.qmd >}}
{{< include _MAYV.qmd >}}
{{< include /_js/_ojs_data.qmd >}}
{{< include /_js/_ojs_family.qmd >}}This page is rendered by Quarto (no _ in front of it). We see the same ingredients as for the family pages:
- the current virus family but also the current virus is set based on the
paramsin the header - the two data parsing libraries are included:
_ojs_data.qmdand_ojs_family.qmd.
The line in the middle inlcude the _MAYV.qmd file which is the file we discussed earlier. The same content is loaded when selecting a virus in the family page as the one we show here in the virus-specific page.
These virus pages are automatically generated using the update_family.js script.
Toolbox

The toolbox is a little bit different in nature, it is complete rendered dynamically based on code. The main page is tooblox/index.qmd and this page performs a few steps:
- load the
_ojs_data.qmdfile in order to load all information for all viruses and the toolbox - Loop over the tools and render them one by one. For every tool, render a table of viruses for which this tool is available.
Some Javascript libraries are used to make this work, but all in all it’s not a difficult page to create.
Grouping
We included a feature to group viruses.
TODO: Extend
Footnotes
Please note that the only difference between
alphaviruses/index.qmdandfiloviruses/index.qmdis this parameter. The rest of the file is generic.↩︎The fact that some files start with an underscore
_MAYV.qmdfor instance is this tells Quarto to not render these as standalone web pages. Generally speaking, we use those files to include in other (rendered).qmdfiles.↩︎